This project aims at investigating current and new hypotheses on the peopling history of East Asia by modern humans.
Project scope
- Did the first Homo sapiens arrive in East Asia from East Africa and the Near East through a unique, southern coastal route, or did they also follow another route north to the Himalayas to occupy Northeast Asia?
- When and from where did the speakers of the Austroasiatic linguistic family expand to achieve their current geographic distribution between India and Southeast Asia?
These and other questions related to human settlement history are addressed through the molecular and bio-statistical analysis of several genes involved in our immunity: the HLA genes.
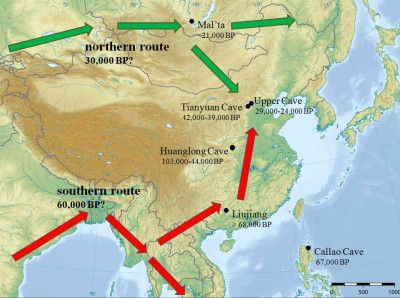
Computer simulation is applied to test the likelihood of the “two-migration routes” model proposed for the peopling of East Asia by modern humans (Di & Sanchez-Mazas 2011; Di, Currat et al. in prep.)
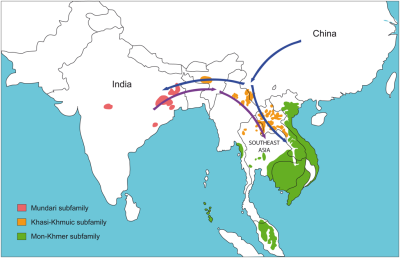
HLA genes, mtDNA, Y-chromosome and classical markers are analysed jointly in the same populations to investigate the genetic history of the austro-asiatic linguistic family (Riccio et al. 2011, Riccio et al. in prep.)
Because HLA genes protect individuals against pathogens but are also associated to a number of infectious and autoimmune diseases, this project also explores the mechanisms underlying the evolution of the HLA genes in human populations:
- Are human populations more resistant to pathogens when exhibiting a higher diversity for their HLA genes?
- Did different HLA genes evolve through different mechanisms, i.e. neutrality vs diversifying selection?
These and other questions related to evolutionary genetics are investigated through the development and application of new computer tools including resampling and simulation algorithms.
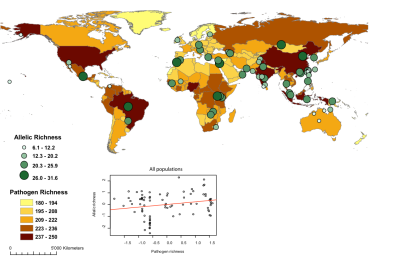
The diversity of HLA genes is partly related to the distribution of pathogen richness in the world (Sanchez-Mazas, Lemaître & Currat 2012, paper presented at the Discussion meeting on Human evolution, migration and history revealed by genetics, immunity and infection of the Royal Society of London).
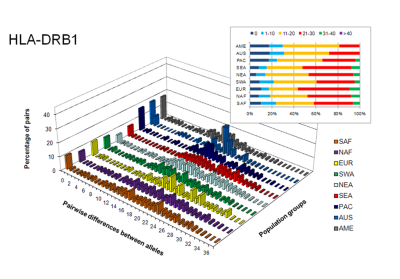
The high number of nucleotide differences between HLA alleles possibly confers higher fitness to the heterozygous individuals against pathogens (Buhler & Sanchez-Mazas 2011).
Project innovations
New population samples
Indian and Vietnamese populations speaking languages of distinct linguistic families (Austroasiatic, Austronesian and Tai-Kadai); Afghans populations speaking either Indo-European or Altaic languages.
New methods in population genetics
Gene[rate], user-friendly computer tools based on resampling algorithms to analyse complex and ambiguous HLA genetic data; Selector, a program based on simulation algorithms to test alternative scenarios of human migrations, including different evolutionary models.
An inter-disciplinary approach
HLA genes are compared to other genetic markers (mtDNA, Y-chromosome, classical markers); genetic data are compared to linguistic and archaeological data.
Partenaires
- Dr Jean-Marie Tiercy (HUG, Geneva)
- Dr Laurent Sagart (CRLAO, Paris)
- Dr Jacques Chiaroni (EFS Marseilles)
- Dr Julie Di Cristofaro (EFS Marseilles)
- Prof. An-Vu-Trieu (Hanoi, Vietnam)
- Prof. Peter Bellwood (School of Archaeology and Anthropology, Australian National University)
- Dr Peter Underhill (Department of Genetics, Stanford University)
Sélection de publications en relation avec le projet
- Sanchez-Mazas, A.; Lemaître, J.-F.; Currat M. (2012) Distinct evolutionary strategies of human leucocyte antigen loci in pathogen-rich environments. Philosophical Transactions of the Royal Society B, Biological Sciences 367 (1590) 830-839 http://archive-ouverte.unige.ch/unige:18732
- Buhler, S.; Sanchez-Mazas, A. (2011) HLA DNA sequence variation among human populations: molecular signatures of demographic and selective events. PLoS One, 6(2):e14643, http://archive-ouverte.unige.ch/unige:14336.
- Di, D.; Sanchez-Mazas, A. (2011) Challenging views on the peopling history of East Asia: the story according to HLA markers. American Journal of Physical Anthropology, 145(1):81–96, http://archive-ouverte.unige.ch/unige:15003.
- Nunes, J.M.; Riccio, M.E.; Tiercy, J-M., Sanchez-Mazas, A. (2011) Allele frequency estimation from ambiguous data: using resampling schema in validating frequency estimates and in selective neutrality testing. Human Biology. 83:437-47, http://archive-ouverte.unige.ch/unige:16925.
- Riccio, M.E.; Nunes, J.M.; Rahal, M.; Kervaire, B.; Sagart, L.; Tiercy, J-M.; Sanchez-Mazas, A. (2011) The Austro-Asiatic Munda population from India and its enigmatic origin: a HLA diversity study. Human Biology, 83:405-35, http://archive-ouverte.unige.ch/unige:16904.
- Sanchez-Mazas, A.; Fernandez-Viña, M.; Middleton, D., Hollenbach, J.A.; Buhler, S.; Di, D.; Rajalingam, R.; Dugoujon J-M.; Mack, S.; Thorsby, E. (2011) Immunogenetics as a tool in anthropological studies. Immunology , 133(2):143–164, http://archive-ouverte.unige.ch/unige:15717.
- Sanchez-Mazas, A.; Di, D.; Riccio, M.E. (2011) A genetic focus on the peopling history of East Asia: critical views. Rice 4:159-169.
- Currat, M.; Poloni, E.S. ; Sanchez-Mazas, A. (2010) Human genetic differentiation across the Strait of Gibraltar. BMC Evolutionary Biology, 10:237-255, http://archive-ouverte.unige.ch/unige:12636.
- Nunes, J.M.; Riccio, M.E.; Buhler, S.; Di, D.; Currat, M.; Ries, F.; Almada, A.J.; Benhamamouch, S.; Benitez, O.; Canossi, A.; Fadhlaoui-Zid, K.; Fischer, G.; Kervaire, B.; Loiseau, P.; de Oliveira, D.C.M.; Papasteriades, C.; Piancatelli, D.; Rahal, M.; Richard, L.; Romero, M.; Rousseau, J.; Spiroski, M.; Sulcebe, G.; Middleton, D.; Tiercy, J.-M.; Sanchez-Mazas, A. (2010) Analysis of the HLA population data (AHPD) submitted to the 15th International Histocompatibility / Immunogenetics Workshop by using the Gene[rate] computer tools accommodating ambiguous data (ahpd project report). Tissue Antigens Published Online: 22 Mar 2010 , http://archive-ouverte.unige.ch/unige:13158.
Selected computer programs related to the project
- Gene[Rate] - computer tools for the analysis of HLA data in the fields of anthropology and disease-association studies.
